Using PDX Like Me
Introduction:
PDX Like Me uses a search language similar to cBioPortal's Onco Query Language to find PDX tumors that match user specified molecular profiles. The profiles can include mutation, expression, and/or copy number aberration criteria.- The input must start with the word CASE followed by and unique identifier.
- Following the line for the case, include one or more lines of search criteria using the format <GeneSymbol>:<search parameter>.
- One criterion per line.
- Gene symbols must conform to official HUGO Gene Nomenclature (HGNC).
- More than one CASE can be submitted at one time.
- Criteria are combined using 'OR'.
Example: | |||||||||||||||||||||
| CASE 1 | |||||||||||||||||||||
|
|
Keywords:
| Data Type | Parameters | Notes |
|---|---|---|
| Mutations | MUT MUT=<protein change> | MUT will return all protein changes MUT=<protein change> will return the specific protein change |
| Copy Number Alterations | AMP DEL NOCNV | LRP:Log Ratio Ploidy = log2(raw copy number / ploidy) AMP is when LRP is greater than 0.5 DEL is when LRP is less than -0.5 NOCNV is for LRP values greater than -0.5 and less than 0.5 |
| Expression | EXP > # EXP < # | # can be a positive or negative integer or decimal value for Z score percentile rank |
| Functional consequence | GOF LOF UNK NONE | GOF = Gain of function LOF = Loss of function UNK = Unknown functional mutation NONE = No functional mutation Functional consequence annotations are derived from the JAX Clinical Knowledgebase (CKB) |
Example Search Results:
Search results are returned as one table per case. Rows correspond to matching models and columns to search criteria. An X in a cell indicates the model matches the criteria.Rows are sorted with the most matches first then by model ID. If selected, CNV and expression values are only shown when matching, clinically relevant variants are shown for all genes where applicable.
CASE 1
| Model ID | KRAS Amp | TP53 MUT=A159V | ALB Del | KIT EXP>1.5 |
| TM00098 invasive ductal carcinoma:Breast | X Log ratio ploidy = 0.65 | Clinically relevant TP53 variant P72R Effect: unknown | X Log ratio ploidy = -1.16 | X Z score percentile rank = 1.89 |
| TM00231 lung squamous cell carcinoma:Lung | X Log ratio ploidy = 1.86 Clinically relevant KRAS variant G12C Effect: loss of function Treatment Approach: MEK inhibitor (Pan),MEK1 Inhibitor | Clinically relevant TP53 variant P72R Effect: unknown | X Log ratio ploidy = -1.5 | Clinically relevant KIT variant M541L Effect: unknown |
| TM01517 adenocarcinoma:Rectum | X Log ratio ploidy = 0.64 Clinically relevant KRAS variant G12C Effect: loss of function Treatment Approach: MEK inhibitor (Pan),MEK1 Inhibitor,MEK2 Inhibitor,PI3K Inhibitor (Pan) | Clinically relevant TP53 variant E286K Effect: loss of function - predicted Treatment Approach: p53 Activator,p53 Gene Therapy | X Log ratio ploidy = -0.57 | Clinically relevant KIT variant V530I Effect: gain of function Treatment Approach: KIT Inhibitor |
| J000094707 rectum adenocarcinoma:Rectum | X Log ratio ploidy = 0.79 Clinically relevant KRAS variant Q61H Effect: loss of function Treatment Approach:PI3K Inhibitor (Pan),PIK3CA inhibitor,RAS Inhibitor (Pan),AZD4785 | Clinically relevant TP53 variant P72R Effect: unknown | X Log ratio ploidy = -0.93 |
Visualization Results
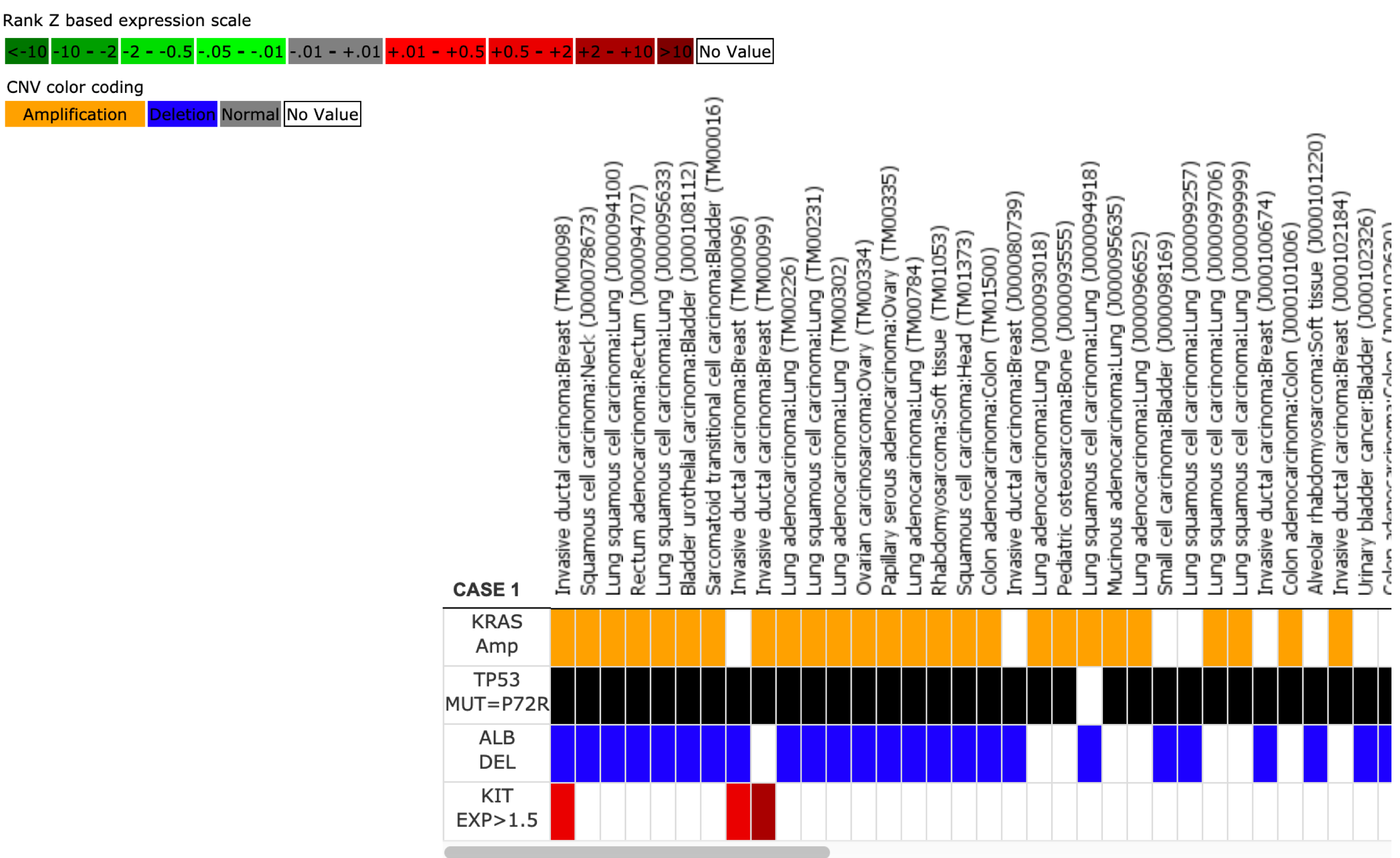
The same results are displayed as a regular search but formatted differently.
Rows display search criteria, columns correspond to matching models.
If multiple cases are used each is displayed in its own tab. Select the tab to see the case results.
Columns can be reordered. Click and hold on the model name, drag to move, click to drop.
Clicking on model names will link to PDX details page for selected model.
If displaying actionable variants, results with an "x" have actionable variants and details will be displayed on mouse-over.
In some cases the tables may render such that the column headers don't align with the rest of the table, usually resizing the browser window width by a small amount will resolve this.